Abstract
The monofunctional platinum(II) complex, phenanthriplatin, acts by blocking transcription, but its regulatory effects on long-noncoding RNAs (lncRNAs) have not been elucidated relative to traditional platinum-based chemotherapeutics, e.g., cisplatin. Here, we treated A549 non-small cell lung cancer and IMR90 lung fibroblast cells for 24 h with either cisplatin, phenanthriplatin or a solvent control, and then performed microarray analysis to identify regulated lncRNAs. RNA22 v2 microRNA software was subsequently used to identify microRNAs (miRNAs) that might be suppressed by the most regulated lncRNAs. We found that miR-25-5p, -30a-3p, -138-5p, -149-3p, -185-5p, -378j, -608, -650, -708-5p, -1253, -1254, -4458, and -4516, were predicted to target the cisplatin upregulated lncRNAs, IMMP2L-1, CBR3-1 and ATAD2B-5, and the phenanthriplatin downregulated lncRNAs, AGO2-1, COX7A1-2 and SLC26A3-1. Then, we used qRT-PCR to measure the expression of miR-25-5p, -378j, -4516 (A549) and miR-149-3p, -608, and -4458 (IMR90) to identify distinct signaling effects associated with cisplatin and phenanthriplatin. The signaling pathways associated with these miRNAs suggests that phenanthriplatin may modulate Wnt/β-catenin and TGF-β signaling through the MAPK/ERK and PTEN/AKT pathways differently than cisplatin. Further, as some of these miRNAs may be subject to dissimilar lncRNA targeting in A549 and IMR90 cells, the monofunctional complex may not cause toxicity in normal lung compared to cancer cells by acting through distinct lncRNA and miRNA networks.
Similar content being viewed by others
Introduction
The platinum(II) complex, cis-diamminedichloroplatinum(II) (cisplatin; Fig. 1), is a bifunctional chemotherapy drug that upon aquation of its two chloride leaving ligands and entry into cancer cell nuclei, typically forms intrastrand crosslinks with DNA causing the nucleotide strands to bend and alteration of gene transcription1,2. As cisplatin chemotherapy is frequently associated with severe side-effects3,4,5,6, there has been considerable interest in identifying alternative platinum complexes that are toxic to cancer cells without causing severe side-effects. The monofunctional platinum(II) complex, cis-[Pt(NH3)2Cl (phenanthridine)]+ (phenanthriplatin; Fig. 1) has similar or superior anticancer efficacy as cisplatin in lung, breast, prostate, testicular, colorectal, glioblastoma and cervical cell lines7,8. Unlike cisplatin, monofunctional compounds have only one chloride leaving ligand and bind to a single strand of DNA without causing DNA to bend7,9. Studies have shown that phenanthriplatin can prevent RNA transcription and has a distinct DNA residue binding profile compared to cisplatin7,10.
Chemical structures of cisplatin and phenanthriplatin. Cisplatin is a bifunctional platinum(II) complex with two chloride leaving ligands. The monofunctional platinum(II) complex, phenanthriplatin, has one chloride leaving ligand, and steric bulk in the form of a heterocyclic phenanthridine ligand placed cis to the platinum coordinating site.
Microarray studies suggest that cisplatin can regulate genes associated with drug resistance, metabolism, cell proliferation, apoptosis, cell adhesion, stress response, cell cycle control and DNA repair11,12,13,14. Long non-coding RNAs (lncRNAs) are RNAs more than 200 nucleotides in length and may interact with protein, chromatin and RNA targets where they can modulate epigenetic, transcriptional, and post-transcriptional function15,16. LncRNA expression can become dysregulated in lung cancer tissue, where their aberrant expression can promote tumor cell growth, apoptosis, invasion, and metastasis15. MicroRNAs (miRNAs) are non-coding RNAs that function post-transcriptionally by binding to messenger RNA (mRNA) to prevent transcription or promote mechanisms that increase mRNA degradation and can regulate tumorigenesis and metastasis16. Further, microarray analysis of lung cancer cells has shown that cisplatin can alter the expression of a lncRNA which acts as a regulatory sponge for a miRNA and modulates chemosensitivity17. Other studies conducted with microarrays in cisplatin-treated cells have shown that lncRNAs are involved in the regulation of cisplatin sensitivity in non-small cell lung cancer (NSCLC)18,19,20. However, the effect of monofunctional platinum(II) complexes on lncRNA expression and their potential regulatory interaction with miRNAs has not yet been characterized.
This study used microarray analysis to investigate whether phenanthriplatin utilizes different lncRNA signaling than cisplatin. The A549 non-small cell lung cancer (NSCLC) cell line is routinely used to evaluate platinum-based gene modulation, and the non-cancer lung fibroblast cell line, IMR90, has been used to assess cisplatin’s effect on gene regulation21,22. Here, we treated A549 and IMR90 cells with the monofunctional complex, cisplatin or a negative media only control. Then, RNA samples were extracted from the cell lines and subjected to microarray analysis. Next, lncRNAs found in the microarray results were bioinformatically analyzed to identify miRNAs with high targeting homology with the lncRNA sequences. qRT-PCR analysis was also used to validate the microarray results and identify miRNAs subject to distinct modulation by the platinum compounds. We found that phenanthriplatin treatment caused up- and downregulation of lncRNAs that were not modulated by cisplatin in either A549 or IMR90 cells. Also, in A549 cells, lncRNAs downregulated by the monofunctional complex have several potential miRNA binding partners associated with Wnt/β-catenin and transforming growth factor-β (TGF-β) signaling that were predicted to also target lncRNAs that were upregulated by cisplatin treatment. This result suggests that cisplatin and phenanthriplatin may utilize distinct lncRNA and miRNA networks to promote anticancer function. Further, we found that phenanthriplatin modulated different lncRNAs in IMR90 cells and that some of the miRNAs predicted to cause cell toxicity during phenanthriplatin treatment in A549 NSCLC cells might promote an opposite effect in IMR90 fibroblast cells.
Results
Microarray and lncRNA qRT-PCR results
Affymetrix microarray analysis was performed on samples taken from cisplatin and phenanthriplatin treated A549 and IMR90 cells to identify regulated lncRNAs. We used a fold change threshold of ≥ 1.5 to identify the most up- and downregulated lncRNAs in A549 cells treated with cisplatin or phenanthriplatin compared to control (Table 1).
Microarray analysis was also used to identify regulated lncRNAs in IMR90 cells treated with cisplatin or phenanthriplatin compared to control samples (Table 2).
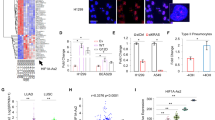
To confirm the microarray data, we also performed qRT-PCR on samples taken from both the A549 and IMR90 cell lines and found that both methods detected similar expression profiles for many of the lncRNAs (Fig. 2). Cisplatin treated A549 samples had the following relative fold expression values (microarray compared to qRT-PCR respectively): PTBP1-2, 1.71–0.63; IMMP2L-1, 1.90–0.99; RIPPLY3-5, 2.20–2.71, which were within approximately one fold change value of one another or less (Fig. 2a). Phenanthriplatin treated A549 samples had very similar relative fold change values for two lncRNAs, TRIM55-1 (2.39–1.82) and RARB-1 (1.82–1.75), while RIPPLY3-5 (2.04–4.83) and PTBP1-2 (2.75–0.33) were within three relative fold change values of each other (Fig. 2a). For cisplatin treated IMR90 samples, relative fold expression values (microarray compared to qRT-PCR respectively) were within approximately one fold change value of one another: MRPS5-1 (1.87–0.71), HYPM-1 (1.30–0.59), NLRP12-1 (1.93–0.75), while several phenanthriplatin treated samples, HYPM-1 (2.90–0.57), FAM237B-2 (3.27–8.06), TMEM243-1 (2.36–0.59) differed by more than a fold change value, with the exception of TRIM55-1 (4.87–5.79), where relative expression was within a fold change value (Fig. 2b).
Validation of long non-coding RNA microarray expression data using qRT-PCR. (a) A549 cell line results for PTBP1-2 (PT), IMMP2L-1 (IM), RIPPLY3-5 (RI), TRIM55-1 (TR), and RARB-1 (RA). (b) IMR90 cell line results for MRPS5-1 (MR), HYPM-1 (HY), NLRP12-1 (NL), FAM237B-2 (FA), TMEM243-1 (TM), and TRIM55-1 (TR). Key: “-M” = microarray (light gray); “-P” = qRT-PCR (dark gray). Error bars represent mean ± standard deviation, N = 3.
Bioinformatics and miRNA qRT-PCR results
RNA22 v2 microRNA software was then used to identify microRNAs predicted to target the three most upregulated lncRNAs in A549 cells after cisplatin treatment. We found that cisplatin upregulated the lncRNAs, ATAD2B-5, CBR3-1, and IMMP2L-1, which may be targeted by several miRNAs with sequence homologies to lncRNAs downregulated by phenanthriplatin, including: AGO2-1, COX7A1-2 and SLC26A3-1 (Table 3; Supplementary Table S1). Also, we found that cisplatin downregulated the lncRNA, lnc-PGPEP1L-3, which is predicted to sponge miR-25-5p, a miRNA with high sequence homology to lnc-MRPL39-10, a lncRNA upregulated by phenanthriplatin (Table 3; Supplementary Table S1). These miRNAs are associated with Wnt/β-catenin and TGF-β signaling, e.g., miR-25-5p, -30a-3p, -138-5p, -149-3p, -185-5p, -378j, -608, -650, -708-5p, -1253, -1254 and -4516 (Table 3)23,24,25,26,27,28,29,30,31,32,33,34,35.
As signaling integrating lncRNAs and miRNAs in IMR90 cells might modulate distinct targets than those in A549 cells, we also identified microRNA targets of the three most upregulated lncRNAs after cisplatin treatment. We found that cisplatin upregulated the lncRNAs, ATAD2B-5, GNG11-3, and MRPS5-1, which may be targeted by miRNAs with sequence homologies to lncRNAs downregulated by phenanthriplatin, including: AGO2-1, ARCN1-1, HYPM-1 (Table 4; Supplementary Table S2). Cisplatin and phenanthriplatin treatment also downregulated the lncRNA, HYPM-1, which may sponge miR-608, and the monofunctional complex upregulated the lncRNA, TMEM243-1, with high sequence homology to miR-608 (Table 4; Supplementary Table S2). As in the A549 results, these miRNAs can potentially act through Wnt/β-catenin and TGF-β signaling (Table 4)23,26,27,29,32,33,34,36.
To determine the expression of miRNA targets of lncRNAs identified in the bioinformatics analysis, we also performed qRT-PCR using locked nucleic acid primers on select miRNAs and found that cisplatin and phenanthriplatin treatment caused different regulation of three miRNAs, miR-25-5p, -378j, and -4516, in the A549 cell line and one miRNA, miR-4458, in the IMR90 cell line (Fig. 3).
Measurement of miRNA expression using qRT-PCR. (a) A549 cell line results for miR-25-5p, -378j and -4516. (b) IMR90 cell line results for miR-149-3p, -608 and -4458. Key: “cis” = cisplatin; “phen” = phenanthriplatin. Error bars represent mean ± standard deviation, N = 3, “*” = p ≤ 0.05; “**” = p ≤ 0.01; “****” = p ≤ 0.0001.
As a final step, we also analyzed how cisplatin and phenanthriplatin regulated lncRNAs might target miRNAs and potentially regulate Wnt/β-catenin and TGFβ signaling in both A549 (Fig. 4) and IMR90 cells (Fig. 5).
Potential modulation of Wnt/β-catenin and TGF-β signaling via cisplatin and phenanthriplatin regulation of lncRNAs and predicted miRNA binding partners in A549 cells. Key: Grey miR text = miRNA associated with downregulated Wnt/β-catenin and TGF-β signaling, Black miR text = miRNA associated with upregulated Wnt/β-catenin and TGF-β signaling, grey open down arrow = downregulated, black open up arrow = upregulated.
Potential modulation of Wnt/β-catenin and TGF-β signaling via cisplatin and phenanthriplatin regulation of lncRNAs and predicted miRNA binding partners in IMR90 cells. Key: Grey miR text = miRNA associated with downregulated Wnt/β-catenin and TGF-β signaling, Black miR text = miRNA associated with upregulated Wnt/β-catenin and TGF-β signaling, grey open down arrow = downregulated, black open up arrow = upregulated.
Discussion
Cisplatin can act through lncRNAs that regulate disease progression in lung cancer by sponging miRNAs, whose interaction with lncRNAs modulates a variety of genes in cancer apoptosis, migration, proliferation and resistance37,38,39,40,41,42. As the role of lncRNAs in the anticancer effect of monofunctional platinum(II) complexes had not yet been assessed, we used microarray analysis to identify regulated lncRNAs in A549 and IMR90 cells after treatment with cisplatin or phenanthriplatin (Tables 1, 2). To validate our microarray results, we performed qRT-PCR on total RNA extracts from cisplatin and phenanthriplatin treated cell preparations and found that the relative fold change expression of regulated lncRNAs calculated via PCR amplification generally corroborated the microarray data (Fig. 2). Next, we applied bioinformatics methods to the microarray results and discovered that a set of miRNAs predicted to target the three most upregulated lncRNAs by cisplatin treatment also had high sequence homology to the three most downregulated lncRNAs, AGO2-1, COX7A1-2 and SLC26A3-1, after phenanthriplatin treatment (Tables 1, 3). Then, we used LNA qRT-PCR to comparatively measure the effect of cisplatin and phenanthriplatin treatment on the expression of several miRNAs that are predicted to be targeted by these lncRNAs (Fig. 3). These results suggest that cisplatin might upregulate a set of lncRNAs that sponge and nullify the gene regulatory effect of these miRNAs; whereas, phenanthriplatin, by reducing the expression of lncRNAs that could prevent the action of the same miRNAs, might promote miRNA gene silencing. The results also suggest that cisplatin and phenanthriplatin treatment might cause distinct regulation of miRNAs, particularly in A549 cells.
Several of the miRNAs detected in our analysis are associated with Wnt/β-catenin signaling. Expression of miR-25-5p may be targeted by two lncRNAs regulated by phenanthriplatin, e.g., lnc-COX7A1-2, that the monofunctional complex reduces, and lnc-MRPL39-10, which phenanthriplatin increases (Table 3). The sponging of miR-25-5p via increased lnc-MRPL39-10 expression could promote anticancer effect through the Wnt pathway as the reduction of this miRNA in A549 cells is associated with decreased cell viability, invasion and migration by decreasing Wnt signaling (Fig. 4)25. Our miRNA qRT-PCR results may corroborate lnc-MRPL39-10 suppression of miR-25-5p as phenanthriplatin treated A549 cells had decreased miR-25-5p expression compared to cisplatin treated samples (Fig. 3a).
As phenanthriplatin reduced the expression of the lncRNAs, AGO2-1, COX7A1-2, and SLC26A3-1 (Table 3), this could promote the action of miRNAs that might be sponged by these lncRNAs. Increased miR-149-3p and miR-185-5p expression downregulates Wnt/β-catenin signaling reducing cancer proliferation in A549 and lung adenocarcinoma cells33,34. Overexpression of miR-608 induced cell death in A549 cells via signaling that integrates the Wnt pathway23. MiR-708-5p expression also suppressed DNA methylation decreasing Wnt/β-catenin signaling and impairing the stemness characteristics of NSCLC cells26. Therefore, phenanthriplatin could act to suppress NSCLC by repressing lncRNAs that regulate miR-25-5p, -185-5p, -608, and -708-5p, through the Wnt/β-catenin pathway (Fig. 4).
However, increasing the expression of some of the miRNAs may also promote cancer through the Wnt/β-catenin pathway. Suppression of miR-378 in A549 cells inactivates Wnt/β-catenin signaling leading to reduced cell proliferation28, and as phenanthriplatin treatment caused reduced miR-378j compared to cisplatin (Fig. 3a), the monofunctional complex may act through this pathway to prevent cancer proliferation. MiR-650 can be upregulated in A549 cells where it promoted proliferation and invasion through the Wnt-1/β‑catenin pathway by acting on inhibitor of growth 429. Increased expression of miR-1253, which directly targets Wnt5A, is associated with the proliferation, migration, and invasion of NSCLC cells27. Similarly, miR-1254 targets the Wnt/β-catenin pathway antagonist, secreted frizzled related protein 1 (SFRP1), and was upregulated in lung cancer tissues and cells promoting proliferation24. Our results suggest that the monofunctional complex might act against NSCLC via Wnt/β-catenin signaling if the miRNAs, miR-149-3p, -185-5p, -608, and -708-5p were not sponged by the phenanthriplatin downregulated lncRNAs, AGO2-1, COX7A1-2 and SLC26A3-1. Nonetheless, increased expression of the miRs, -25-5p, -650, -1253, and -1254, when these lncRNAs are suppressed by the monofunctional complex, could promote cancer by modulating the Wnt/β-catenin pathway possibly in conjunction with other pathway signaling (Fig. 4).
LncRNAs are also able to act in NSCLC through the TGF-β pathway and integrated MAPK/ERK and PI3K/AKT signaling43,44. We identified several miRNAs that might integrate TGF-β signaling components through lncRNAs regulated by phenanthriplatin (Fig. 4). Overexpression of miR-608 induced cell death in A549 cells through the MAPK/ERK pathway23. As the three lncRNAs, AGO2-1, COX7A1-2, and SLC26A3-1, which are predicted to sponge miR-608, were downregulated by the monofunctional complex (Table 3), we would expect that expression of this miRNA would lead to NSCLC cell death via ERK signaling. However, suppression of AGO2-1, COX7A1-2, and SLC26A3-1, could also have an opposite effect in NSCLC cells by upregulating the miRs, -25-5p, -30a-3p and -378, which act via MAPK/ERK, and are projected targets of these lncRNAs (Table 3), and have reduced anti-cancer efficacy when their expression is increased25,28,30. We found that phenanthriplatin treatment downregulated miR-378j more than cisplatin (Fig. 3a), which suggests that the monofunctional complex may signal via the TGF-β/MAPK/ERK pathway to promote anti-cancer effect. As miR-25-5p may be sponged via phenanthriplatin-mediated upregulation of lnc-MRPL39-10 (Table 3), this miRNA could be downregulated in A549 cells, which is associated with anti-cancer effect via PI3K signaling25. Reduction of AGO2-1, COX7A1-2, and SLC26A3-1 by phenanthriplatin would also be expected to lead to increased miR-138-5p and -608, whose expression modulates PTEN/PI3K/AKT signaling and is associated in A549 cells with anticancer effect23,31,45,46,47. In retinoblastoma tumors, increased miR-4516 expression promotes tumor growth through the PTEN/AKT pathway32. As phenanthriplatin treatment caused increased miR-4516 expression in A549 cells compared to cisplatin (Fig. 3a), the monofunctional complex may be able to promote PTEN/AKT signaling more than its bifunctional counterpart.
We also examined whether cisplatin and phenanthriplatin might regulate Wnt/β-catenin and TGF-β signaling differently in IMR90 compared to A549 cells through lncRNA regulation of miRNAs. Interestingly, a majority of the miRNAs, e.g., miR-149-3p, -185-5p, -608, -650, -708-5p, and -4516, identified as potential targets of lncRNAs in A549 cells, were predicted to be regulated identically in IMR90 cells albeit by different lncRNAs (Table 4, Fig. 5). Specifically, cisplatin upregulates lncRNAs, e.g., ATAD2B-5, GNG11-3, and MRPS5-1, that collectively should sponge all of these miRNAs, and phenanthriplatin downregulates lncRNAs, e.g., AGO2-1, ARCN1-1, and HYPM-1, that should allow expression of these miRNAs. Cisplatin also downregulates HYPM-1, which should allow expression of miR-608; whereas, the monofunctional complex upregulates TMEM243-1, a lncRNA with high sequence homology to this miRNA. Therefore, regulation of Wnt/β-catenin and TGF-β signaling through miR-149-3p, -185-5p, -608, -650, -708-5p, and -4516 would be predicted to be similar in both IMR90 and A549 cells. However, it is possible that simultaneous upregulation of multiple lncRNAs, e.g., ATAD2B-5 and MRPS5-1 in cisplatin treated IMR90 cells, and downregulation of multiple lncRNAs, e.g., AGO2-1, ARCN1-1, and HYPM-1 by phenanthriplatin, could synergistically promote or prevent respectively the action of other lncRNAs and might alter Wnt/β-catenin and TGF-β signaling differently than would occur from the effect of these lncRNAs and miRNAs alone. When we measured miR-149-3p and -608 expression in the IMR90 cell line, we found that cisplatin and phenanthriplatin did not exhibit distinct regulation of either miRNA (Fig. 3b). Thus, it is possible that the action of specific lncRNAs to modulate miRNA expression, and Wnt/β-catenin and TGF-β signaling, could be masked by the effect of larger combinations of up- and downregulated lncRNAs acting together.
Further, we also identified a potential miRNA target, miR-4458, unique to IMR90 cells. Cisplatin treatment upregulated lncRNA, ATAD2B-5, which might sponge miR-4458, while phenanthriplatin downregulated both AGO2-1 and HYPM-1, lncRNAs with targeting homology to this miRNA (Table 4). Interestingly, cisplatin treatment in IMR90 cells caused increased miR-4458 expression compared to phenanthriplatin (Fig. 3b), which could mean that increased ATAD2B-5 is less effective as a suppressor of this miRNA than the effect of decreasing either AGO2-1 or HYPM-1. As increased expression of miR-4458 prevents proliferation and migration in NSCLC cell lines via the PI3K/AKT pathway36, cisplatin treatment might suppress miR-4458 with ATAD2B-5 allowing cancer progression, while phenanthriplatin, by decreasing the expression of AGO2-1 and HYPM-1, could allow miR-4458 expression through modulating the PI3K/AKT pathway to promote anti-cancer activity.
As in A549 cells, phenanthriplatin regulated a distinct set of lncRNAs compared to cisplatin in IMR90 cells (Tables 1, 2) and might modulate Wnt/β-catenin and TGF-β signaling differently in normal lung fibroblast cells than in lung cancer cells (Figs. 4, 5). In A549 cells, miR-25-5p expression may be sponged by upregulated lnc-MRPL39-10, but we found in IMR90 cells that this miRNA would be potentially decoyed by lnc-HYPM-1, which is downregulated by phenanthriplatin (Tables 2, 3, 4). The absence of suppression of miR-25-5p by lnc-HYPM-1 in IMR90 cells could promote cell proliferation and viability via Wnt/β-catenin and/or TGF-β signaling integrating MAPK/ERK as found in A549 cells25. Similarly, miR-138-5p and miR-608 are potentially not subject to sponging in A549 cells treated with the monofunctional complex as their lncRNA targets are downregulated, but in IMR90 cells, these miRNA could be sponged by upregulated lnc-TMEM243-1 (Tables 2, 3, 4). Therefore, reduced expression of miR-138-5p and -608 due to TMEM243-1 decoying in IMR90 cells could lead to reduced cytotoxicity via TGF-β signaling (miR-138-5p) or Wnt/β-catenin and TGF-β signaling (miR-608) as suggested by the effects of suppressing these miRNAs in A549 cells23,46,47,48,49.
Materials and methods
Cell culture and treatment
The A549 and IMR90 cell lines were obtained from ATCC (Manassas, VA). A549 cells were cultured in F12K media with 10% FBS and 1% penicillin/streptomycin supplementation. IMR90 cells were cultured in Eagle's Minimum Essential Medium (EMEM) with 10% FBS and 1% penicillin/streptomycin supplementation. We seeded 6-well dishes with 3 × 105 cells per well and placed them in an incubator (37 °C, 5% CO2) for 24 h. Sets of three wells each were treated with either a negative control (media only), 5 µM cisplatin (Sigma-Aldrich, St. Louis, MO), or 5 µM phenanthriplatin (synthesized by Dr. Kevin M. Williams, Western Kentucky University Department of Chemistry, Bowling Green, KY according to a published protocol8), a platinum concentration that efficiently targets the nucleus at 24 h8. Then, the dishes were placed back into the incubator for 24 h after which they were prepared for RNA isolation.
RNA isolation
The Qiagen RNeasy kit (Hilden, Germany) was used to isolate RNA samples from control and platinum complex-treated dishes per kit instructions. Media was aspirated out of the wells followed by washing (1×) with phosphate buffered saline (PBS). The PBS was removed, and Buffer RLT Plus with β-mercaptoethanol was introduced into each well and homogenate was then transferred to a microcentrifuge tube. Each tube was vortexed and the lysate was transferred to a Gdna Eliminator spin column and centrifuged. Flow-through was collected and 70% ethanol was added. The samples were then transferred to an RNeasy spin column placed in a collection tube. Spin columns were centrifuged and the flow-through was discarded. Buffer RW1 was added to each spin column, followed by centrifugation. Flow-through was discarded and Buffer RPE with ethanol was added followed by centrifugation. The flow-through was again discarded and Buffer RPE with ethanol was then added followed by centrifugation. The spin column was then placed in a collection tube, centrifuged, and then placed in a new collection tube. Two elutions using RNase-free water were performed with centrifugation. All RNA samples were stored at − 80 °C for subsequent analysis.
Microarray assay
RNA transcription profiles from experimental and control samples were created using a microarray assay. RNA samples were analyzed using a NanoDrop 2000 (Thermo Fisher, Waltham, MA) and quality was assessed with an Agilent 2100 Bioanalyzer (Agilent, Wilmington, DE). RNA integrity was expressed as RNA integrity number (RIN) values and only samples with values greater than 10 were used. The Low RNA Input Linear Amplification kit with one-color (Agilent, Wilmington, DE) was used to produce fluorescent cRNA. Labeled cRNA was then purified using the RNeasy Mini Elute kit (Qiagen, Valencia, CA). Fluorescent cRNA samples were then hybridized onto Whole Human Genome 4 × 44 K microarrays GeneChips (Affymetrix, Santa Clara, CA). Four replicates were performed with array hybridization conducted for 17 h at 65 °C. Slides were then scanned with an Agilent microarray scanner (G2565BA) using Feature Extraction software (v 9.5.1, Agilent). GeneSpring GX 10.0.2 software (Agilent Technologies) was used for statistical analysis, background correction, normalization and summary of expression measure.
Quantitative real-time and locked nucleic acid polymerase chain reaction analysis
Microarray results were additionally validated using quantitative real time-polymerase chain reaction (qRT-PCR) on RNA samples also used for the microarrays. Custom probes were designed and specificity checked using the National Institutes of Health Primer-BLAST tool and obtained from Integrated DNA Technologies (Coralville, IA). Samples were placed in an Agilent Technologies Stratagene Mx3000P real-time PCR System (Santa Clara, CA) with MxPro software with thermal cycling at 95 °C for 10 min, followed by 40 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. Additionally, RNA samples were analyzed using locked nucleic acid PCR (LNA) miRNA to measure the expression of miRNA targets (A549: miR-25-5p, -378j, -4516; IMR90: miR-149-3p, -608, -4458). Custom LNA probes were obtained from Qiagen (Hilden, Germany). Samples were analyzed with the Mx3000P and MxPro software as per the qRT-PCR experiments except that thermal cycling was performed at 95 °C for 2 min (initial activation), followed by 40 cycles of denaturing at 95 °C for 10 s, and annealing/extension at 56 °C for 1 min. Three independent reactions were performed for each qRT-PCR and LNA PCR experiment with three technical replicates prepared in each. Quantitative data for qRT-PCR and LNA PCR experiments were normalized to the rRNA 5S gene.
Data and statistical analysis
Long noncoding RNA transcript IDs identified by the microarray assay were sequence characterized using the LNCipedia database (https://lncipedia.org/50. Identification of miRNA targeting of lncRNAs used the RNA22 v2 microRNA target detection software (https://cm.jefferson.edu/rna22/Interactive/51 with p ≤ 0.05 as the standard (except that for GNG11-3, p ≤ 0.0581). Microarray data was statistically analyzed using GeneSpring GX 10.0.2 software (Agilent Technologies) with p ≤ 0.05 as the standard. qRT-PCR and LNA PCR data were analyzed using the Livak equation to determine relative fold values and standard deviation fold change was calculated using the formula: s = ((s1)2 + (s2)2)1/2. GraphPad PRISM (version 9.0.0; La Jolla, CA) was used for all other statistical analysis with p ≤ 0.05 as the standard.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Cepeda, V. et al. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med. Chem. 7, 3–18. https://doi.org/10.2174/187152007779314044 (2007).
Dasari, S. & Tchounwou, P. B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 740, 364–378. https://doi.org/10.1016/j.ejphar.2014.07.025 (2014).
Miller, R. P., Tadagavadi, R. K., Ramesh, G. & Reeves, W. B. Mechanisms of Cisplatin nephrotoxicity. Toxins (Basel) 2, 2490–2518. https://doi.org/10.3390/toxins2112490 (2010).
Salehi, P. et al. Attenuation of cisplatin ototoxicity by otoprotective effects of nanoencapsulated curcumin and dexamethasone in a guinea pig model. Otol. Neurotol. 35, 1131–1139. https://doi.org/10.1097/MAO.0000000000000403 (2014).
Karasawa, T. & Steyger, P. S. An integrated view of cisplatin-induced nephrotoxicity and ototoxicity. Toxicol. Lett. 237, 219–227. https://doi.org/10.1016/j.toxlet.2015.06.012 (2015).
Santos, N. A. G. D., Ferreira, R. S. & Santos, A. C. D. Overview of cisplatin-induced neurotoxicity and ototoxicity, and the protective agents. Food Chem. Toxicol. 136, 111079. https://doi.org/10.1016/j.fct.2019.111079 (2020).
Park, G. Y., Wilson, J. J., Song, Y. & Lippard, S. J. Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. USA 109, 11987–11992. https://doi.org/10.1073/pnas.1207670109 (2012).
Monroe, J. D., Hruska, H. L., Ruggles, H. K., Williams, K. M. & Smith, M. E. Anti-cancer characteristics and ototoxicity of platinum(II) amine complexes with only one leaving ligand. PLoS ONE 13, e0192505. https://doi.org/10.1371/journal.pone.0192505 (2018).
Johnstone, T. C., Park, G. Y. & Lippard, S. J. Understanding and improving platinum anticancer drugs—Phenanthriplatin. Anticancer Res. 34, 471–476 (2014).
Riddell, I. A., Johnstone, T. C., Park, G. Y. & Lippard, S. J. Nucleotide binding preference of the monofunctional platinum anticancer-agent phenanthriplatin. Chemistry 22, 7574–7581. https://doi.org/10.1002/chem.201600236 (2016).
Yamano, Y. et al. Identification of cisplatin-resistance related genes in head and neck squamous cell carcinoma. Int. J. Cancer 126, 437–449. https://doi.org/10.1002/ijc.24704 (2010).
Carminati, P. O. et al. Alterations in gene expression profiles correlated with cisplatin cytotoxicity in the glioma U343 cell line. Genet. Mol. Biol. 33, 159–168. https://doi.org/10.1590/S1415-47572010005000013 (2010).
Motamedian, E., Ghavami, G. & Sardari, S. Investigation on metabolism of cisplatin resistant ovarian cancer using a genome scale metabolic model and microarray data. Iran J. Basic Med. Sci. 18, 267–276 (2015).
Wang, S. et al. Cisplatin suppresses the growth and proliferation of breast and cervical cancer cell lines by inhibiting integrin β5-mediated glycolysis. Am. J. Cancer Res. 6, 1108–1117 (2016).
Ginn, L., Shi, L., Montagna, M. & Garofalo, M. LncRNAs in non-small-cell lung cancer. Noncoding RNA 6, E25. https://doi.org/10.3390/ncrna6030025 (2020).
Santos, R. M., Moreno, C. & Zhang, W. C. Non-coding RNAs in lung tumor initiation and progression. Int. J. Mol. Sci. 21, 2774. https://doi.org/10.3390/ijms21082774 (2020).
Zuo, W., Zhang, W., Xu, F., Zhou, J. & Bai, W. Long non-coding RNA LINC00485 acts as a microRNA-195 sponge to regulate the chemotherapy sensitivity of lung adenocarcinoma cells to cisplatin by regulating CHEK1. Cancer Cell Int. 19, 240. https://doi.org/10.1186/s12935-019-0934-7 (2019).
Liu, J. et al. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS ONE 10, e0114586. https://doi.org/10.1371/journal.pone.0114586 (2015).
Hu, L. et al. Aberrant long noncoding RNAs expression profiles affect cisplatin resistance in lung adenocarcinoma. Biomed. Res. Int. 2017, 7498151. https://doi.org/10.1155/2017/7498151 (2017).
Ye, Y. et al. Long non-coding RNA SPRY4-IT1 REVERSES CISPLATIN resistance by downregulating MPZL-1 via suppressing EMT in NSCLC. Onco. Targets Ther. 13, 2783–2793. https://doi.org/10.2147/OTT.S232769 (2020).
Viniegra, J. G. et al. Modulation of PI3K/Akt pathway by E1a mediates sensitivity to cisplatin. Oncogene 21, 7131–7136. https://doi.org/10.1038/sj.onc.1205934 (2002).
Zhang, Y. et al. Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Sci. 111, 637–646. https://doi.org/10.1016/j.prp.2018.10.010 (2020).
Othman, N., In, L. L., Harikrishna, J. A. & Hasima, N. Bcl-xL silencing induces alterations in hsa-miR-608 expression and subsequent cell death in A549 and SK-LU1 human lung adenocarcinoma cells. PLoS ONE 8, e81735. https://doi.org/10.1371/journal.pone.0081735 (2013).
Li, H., Yang, T., Shang, D. & Sun, Z. miR-1254 promotes lung cancer cell proliferation by targeting SFRP1. Biomed. Pharmacother. 92, 913–918. https://doi.org/10.1016/j.biopha.2017.05.116 (2017).
Zhan, J. W. et al. Integrated microRNA and gene expression profiling reveals the crucial miRNAs in curcumin anti-lung cancer cell invasion. Thorac. Cancer 8, 461–470. https://doi.org/10.1111/1759-7714 (2017).
Liu, T., Wu, X., Chen, T., Luo, Z. & Hu, X. Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell-like phenotypes through repressing Wnt/β-catenin signaling. Clin. Cancer Res. 24, 1748–1760. https://doi.org/10.1158/1078-0432.CCR-17-1169 (2018).
Liu, M. et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 9, 189. https://doi.org/10.1038/s41419-017-0218-x (2018).
Gao, S., Yu, Y., Liu, L., Meng, J. & Li, G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 233, 116692. https://doi.org/10.1016/j.lfs.2019.116692 (2019).
Tang, X. et al. miR-650 promotes non-small cell lung cancer cell proliferation and invasion by targeting ING4 through Wnt-1/β-catenin pathway. Oncol. Lett. 18, 4621–4628. https://doi.org/10.3892/ol.2019.10805 (2019).
Wei, D., Yu, G. & Zhao, Y. MicroRNA-30a-3p inhibits the progression of lung cancer via the PI3K/AKT by targeting DNA methyltransferase 3a. Onco. Targets Ther. 12, 7015–7024. https://doi.org/10.2147/OTT.S213583 (2019).
Zhu, D. et al. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol. Res. Pract. 215, 861–872. https://doi.org/10.1016/j.prp.2019.01.029 (2019).
Hao, B. et al. miR-4516 inhibits the apoptosis of RB tumor cells by targeting the PTEN/AKT signaling pathway. Exp. Eye Res. 200, 108224. https://doi.org/10.1016/j.exer.2020.108224 (2020).
Jiang, Z. et al. Maimendong and Qianjinweijing Tang (Jin formula) suppresses lung cancer by regulation of miR-149-3p. J. Ethnopharmacol. 258, 112836. https://doi.org/10.1016/j.jep.2020.112836 (2020).
Li, O. et al. Circle RNA FOXP1 promotes cell proliferation in lung cancer by regulating miR-185-5p/Wnt1 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 6767–6778. https://doi.org/10.26355/eurrev_202006_21665 (2020).
Li, L. et al. CircRNA hsa_circ_0087862 acts as an oncogene in non-small cell lung cancer by targeting miR-1253/RAB3D axis. Onco. Targets Ther. 13, 2873–2886. https://doi.org/10.2147/OTT.S243533 (2020).
Ma, Y., Li, X., Chen, S., Du, B. & Li, Y. MicroRNA-4458 suppresses migration and epithelial-mesenchymal transition via targeting HMGA1 in non-small-cell lung cancer cells. Cancer Manag. Res. 11, 637–649. https://doi.org/10.2147/CMAR.S185117 (2019).
Leon, L. M. et al. The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA contributes to an aggressive phenotype in lung adenocarcinoma through regulation of oxidative stress. Oncogene 38, 7146–7165. https://doi.org/10.1038/s41388-019-0935-y (2019).
Ju, L., Han, M., Li, X. & Zhao, C. MicroRNA signature of lung adenocarcinoma with EGFR exon 19 deletion. MicroRNA signature of lung adenocarcinoma with EGFR Exon 19 deletion. J. Cancer 8, 1311–1318. https://doi.org/10.7150/jca.17817 (2017).
Wang, H., Wang, L., Zhang, S., Xu, Z. & Zhang, G. Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. Biomed. Pharmacother. 127, 110214. https://doi.org/10.1016/j.biopha.2020.110214 (2020).
Wu, Y., Cheng, K., Liang, W. & Wang, X. lncRNA RPPH1 promotes non-small cell lung cancer progression through the miR-326/WNT2B axis. Oncol. Lett. 20, 105. https://doi.org/10.3892/ol.2020.11966 (2020).
Zeng, Z. et al. LncRNA FOXD3-AS1 promoted chemo-resistance of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. Cancer Cell Int. 20, 350. https://doi.org/10.1186/s12935-020-01402-9 (2020).
Zhan, Y. et al. Knockdown of long non-coding RNA HOTAIR suppresses cisplatin resistance, cell proliferation, migration and invasion of DDP-resistant NSCLC cells by targeting miR-149-5p/doublecortin-like kinase 1 axis. Cancer Manag. Res. 12, 7725–7737. https://doi.org/10.2147/CMAR.S246299 (2020).
Xu, C. C. et al. Effects of TGF-β signaling blockade on human A549 lung adenocarcinoma cell lines. Mol. Med. Rep. 4, 1007–1015. https://doi.org/10.3892/mmr.2011.530 (2011).
Wang, C. J. et al. MicroRNA-31/184 is involved in transforming growth factor-β-induced apoptosis in A549 human alveolar adenocarcinoma cells. Life Sci. 242, 117205. https://doi.org/10.1016/j.lfs.2019.117205 (2020).
Karreth, F. A. et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395. https://doi.org/10.1016/j.cell.2011.09.032 (2011).
Pan, X., Chen, Y., Shen, Y. & Tantai, J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 10, 429. https://doi.org/10.1038/s41419-019-1660-8 (2019).
Song, N. et al. MicroRNA-138-5p suppresses non-small cell lung cancer cells by targeting PD-L1/PD-1 to regulate tumor microenvironment. Front. Cell Dev. Biol. 8, 540. https://doi.org/10.3389/fcell.2020.00540 (2020).
Guo, L., Chen, Y., Luo, J., Zheng, J. & Shao, G. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio 9, 437–445. https://doi.org/10.1002/2211-5463.12597 (2019).
Wang, Y. et al. MicroRNA-608 sensitizes non-small cell lung cancer cells to cisplatin by targeting TEAD2. Mol. Med. Rep. 20, 3519–3526. https://doi.org/10.3892/mmr.2019.10616 (2019).
Volders, P. J. et al. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 47, D135–D139. https://doi.org/10.1093/nar/gky103 (2019).
Miranda, K. C. et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217. https://doi.org/10.1016/j.cell.2006.07.031 (2006).
Acknowledgements
We would like to thank both Sabine Waigel and Vennila Arumugam of the University of Louisville Genomics Core Facility for performing the microarray and technical assistance and Dr. Kevin Williams of Western Kentucky University for providing the phenanthriplatin.
Funding
This work was supported by the Kentucky Biomedical Research Infrastructure Network and INBRE (KBRIN) Grant ULRF13-1493C-01. Part of this work was performed with assistance of the University of Louisville Genomics Facility, which is supported by NIH P20GM103436 (KY IDeA Networks of Biomedical Research Excellence), the J. G. Brown Cancer Center, and user fees. Additional funding was provided through NIH Grants T1 R15 CA188890-01A1 (Michael Smith, PI), P20 GM104357 (Yann Gibert, PI), and the Intramural Research Support Program of The University of Mississippi Medical Center.
Author information
Authors and Affiliations
Contributions
J.D.M. and M.E.S. conceptualized the project; J.D.M., S.E.M., E.N.I., and M.E.S. validated the data; J.D.M., S.E.M., and E.N.I. performed formal data analysis; J.D.M., S.E.M., E.N.I., and K.E.L. performed the experiments; M.D.H., M.E.S., and Y.G. provided laboratory resources; J.D.M., S.E.M., and E.N.I. performed data curation; J.D.M. wrote the original draft; J.D.M., S.E.M., E.N.I., K.E.L., M.D.H., M.E.S., and Y.G. revised and edited the manuscript; J.D.M. created the figures and tables; J.D.M., M.D.H., M.E.S., and Y.G. supervised the project; M.E.S. and Y.G. provided overall project administration, M.E.S. and Y.G. were responsible for funding acquisition. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Monroe, J.D., Moolani, S.A., Irihamye, E.N. et al. Cisplatin and phenanthriplatin modulate long-noncoding RNA expression in A549 and IMR90 cells revealing regulation of microRNAs, Wnt/β-catenin and TGF-β signaling. Sci Rep 11, 10408 (2021). https://doi.org/10.1038/s41598-021-89911-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89911-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.