Abstract
Coronavirus disease 2019 (COVID-19) was first diagnosed in Scotland on 1 March 2020. During the first month of the outbreak, 2,641 cases of COVID-19 led to 1,832 hospital admissions, 207 intensive care admissions and 126 deaths. We aimed to identify the source and number of introductions of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into Scotland using a combined phylogenetic and epidemiological approach. Sequencing of 1,314 SARS-CoV-2 viral genomes from available patient samples enabled us to estimate that SARS-CoV-2 was introduced to Scotland on at least 283 occasions during February and March 2020. Epidemiological analysis confirmed that early introductions of SARS-CoV-2 originated from mainland Europe (the majority from Italy and Spain). We identified subsequent early outbreaks in the community, within healthcare facilities and at an international conference. Community transmission occurred after 2 March, 3 weeks before control measures were introduced. Earlier travel restrictions or quarantine measures, both locally and internationally, would have reduced the number of COVID-19 cases in Scotland. The risk of multiple reintroduction events in future waves of infection remains high in the absence of population immunity.
Similar content being viewed by others
Main
The pandemic virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread rapidly throughout the world following its emergence in Wuhan, China in December 20191,2,3. SARS-CoV-2 is a highly transmissible Betacoronavirus related to the first SARS virus4. It causes the clinical syndrome coronavirus disease 2019 (COVID-19), which is characterized by nonspecific respiratory or gastrointestinal viral symptoms and anosmia. In severe cases, acute respiratory distress syndrome, cardiovascular disease, neurological manifestations, thrombosis and renal failure may occur5,6. A rare Kawasaki-like disease has been described in children7. Despite the mobilization of substantial resources to contain the outbreak, COVID-19 was declared a Public Health Emergency of International Concern by the World Health Organization (WHO) on 30 January 2020 and a pandemic on 11 March 20208,9. Many countries are now responding to large outbreaks triggering unprecedented social and economic disruption and challenges to local healthcare systems.
The WHO has declared a Public Health Emergency of International Concern on five occasions since 2009, all as a result of RNA virus outbreaks (H1N1 influenza, Zika virus, polio and Ebola). Pathogen genomic sequencing is now established as a core component of the modern epidemiological response to such outbreaks, driven by modern nucleic acid sequencing technologies that can rapidly yield entire pathogen genomes from clinical samples10. The integration of viral genomic data with spatial, temporal and other metadata in a genomic epidemiology framework has allowed enhanced inference of the origin and transmission dynamics of disease outbreaks11,12,13. Such an approach is particularly applicable to RNA viruses, as their relatively low-fidelity replication cycle generates mutations in the viral genome at a rate observable over the rapid time scale of an outbreak14.
In this study, we sequenced laboratory-confirmed cases of COVID-19 in Scotland and analysed them alongside available international data, in order to estimate the number of introduction events and early spread of SARS-CoV-2 in the country. During the sampling period, 2,641 positive cases of COVID-19 were detected (https://www.opendata.nhs.scot and https://statistics.gov.scot), associated with 1,832 hospital admissions, 207 intensive care admissions and 126 deaths. By applying a genomic epidemiology approach to the data, we demonstrate that the outbreak was the result of multiple separate introductions of the virus associated with international travel, and that community transmission was quickly established in Scotland, well before the introduction of lockdown countermeasures on 23 March 2020.
Results
Multiple introductions of SARS-CoV-2 in Scotland
A total of 1,314 SARS-CoV-2 genomes were generated with >90% genome coverage, representing 49% of laboratory-confirmed Scottish COVID-19 cases (Supplementary Data 1). The median Ct value was 28.92 (interquartile range (IQR) = 25.81–32.06). Of 1,314 individuals, 976 (74%) reported no travel and 114 (9%) reported travel outside Scotland in the 2 weeks preceding the onset of symptoms. No travel history was available for 224 (17%) of individuals. Countries visited included Italy (n = 41), Spain (n = 28), Austria (n = 6), Switzerland (n = 4), France (n = 4), England (n = 9), Wales (n = 1), Germany (n = 1), the Netherlands (n = 1), Ireland (n = 1), Poland (n = 1), Cyprus (n = 1), Turkey (n = 1), Argentina (n = 1), Egypt (n = 1), Tunisia (n = 1), Canada (n = 1), the United States (n = 2) and Thailand (n = 1). Seven individuals returned from Caribbean cruise holidays. Samples were drawn from all regions of Scotland, with the exception of the Orkney Islands, in which only three cases were detected during the study period (Extended Data Fig. 1).
The first case of COVID-19 in Scotland (CVR01, where ‘CVR’ represents analysis at the Medical Research Council (MRC)–University of Glasgow Centre for Virus Research) was in a 51-year-old male from Tayside with mild respiratory symptoms who was tested on 28 February 2020 (and reported positive on 1 March 2020). He had returned from Italy after attending a rugby match 9 d earlier15. The first confirmed case in an individual who had not travelled (CVR02) occurred 3 d later, on 2 March 2020. Reflecting the change from returning travellers to an older disease-susceptible demographic, the median age of cases increased from 44 years (IQR = 32–51 years) in the first week of the epidemic to 62 years (IQR = 47–76 years) in the fourth week, as infections moved from travel-associated to local community transmission (Fig. 1; Kruskal–Wallis test; P < 0.001). A large proportion of sampled cases were healthcare workers and this increased from 5.9% during week 1 to 13.9% by week 4 of the outbreak (Extended Data Fig. 2).
a, Histogram of daily cases stratified by confirmed travel history. b, Ages of positive cases referred for diagnostic testing to the West of Scotland Specialist Virology Centre by week of the outbreak in Scotland. Centre bars represent the median, box edges the IQR, vertical lines the range date, and points the outliers. The median age increased from 44 years in week 1 (n = 12; IQR = 18.8) to 50 years in week 2 (n = 52; IQR = 25.8), 61 years in week 3 (n = 183; IQR = 30.5) and 62 years in week 4 (n = 516; IQR = 29). Significance was determined by Kruskal–Wallis rank-sum test (H = 27.47; d.f. = 3; P < 0.001). c, Spatial distribution and associated travel history over the first 4 weeks of the outbreak. Testing of community cases ceased on day 1 of week 3 (14 March) and lockdown occurred on day 2 of week 4 (23 March).
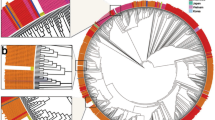
To determine the relationship of the viruses identified in Scotland to global SARS-CoV-2 variants, we inferred an evolutionary tree using viruses sequenced in this study against all complete genomes available from GISAID sampled before 31 March 2020 (GISAID = 9,091; UK-ENG (United Kingdom - England) = 7,801; UK-SCT (United Kingdom - Scotland) = 1,314; UK-WLS (United Kingdom - Wales) = 965; UK-NIR (United Kingdom - Northern Ireland) = 199) (Fig. 2 and Supplementary Data 1).
Known travel histories are indicated (see legend). Global sequence data were available from GISAID. Non-Scottish sequences were subsampled for presentation purposes by keeping a single sequence per date, country or lineage, reducing the total number of non-Scottish sequences from 17,578 to 5,389. The scale bar indicates the number of substitutions per nucleotide site.
While overall limited variability in the genome was observed, in keeping with the lower evolutionary rate of coronaviruses compared with other RNA viruses and the recent introduction of the virus into the human population, the strains introduced into Scotland were diverse, representing much of the global distribution of lineages, with the majority fitting within lineage B16 (Fig. 2). Lineages A.2 (n = 53) and A.5 (n = 22) were the most prevalent A lineages; both are prevalent in Spain (47% of global A.2 and 56% of A.5 lineage sequences on GISAID derive from Spain). However, low sequencing coverage from some regions of the world (for example, China) is likely to have limited this assessment. Scottish sequences occurred throughout the B lineage, with the largest number of sequences (n = 287) found within the B.1 lineage. Most Scottish lineages have also been detected in other parts of the United Kingdom, except for the marked absence of lineage B.1.13 (UK sequences = 165, but 0 in Scotland). An increase in the number of sequences within the B.1, B.1.1, B.2 and B lineages coincided with the number of returning travellers from Italy and other parts of Europe in the first half of March, while the second half of March showed a rise in sequences within the A.2 lineage, coinciding with returning travellers from Spain and other parts of the United Kingdom (Extended Data Fig. 3).
The 1,314 Scottish genomes displayed an average of 3.6 non-synonymous and 1.9 synonymous nucleotide substitutions compared with Wuhan-Hu-1 (Fig. 3). Two common amino acid substitutions (p.Asp614Gly and p.Pro323Leu) were observed in spike and nsp12, respectively, and increased in prevalence over the first month of the epidemic (Fig. 3a–c). Another spike replacement, p.Asn439Lys (reported by the GISAID receptor binding surveillance application CoVsurver; https://www.gisaid.org/covsurver) occurred in addition to p.Asp614Gly in a Scottish cluster of 12 sequences. p.Asp614Gly has been hypothesized to be associated with increased transmissibility and escape from neutralization17, while p.Asn439Lys occurs at a predicted angiotensin-converting enzyme 2 binding site. The Gly614 spike variant was introduced into Scotland an estimated 191 times and was the predominant variant sampled in March, with 807 sequences. In comparison, the Asp614 variant was introduced 91 times, with 504 sequences. Three sequences lacked sufficient coverage at residue 614 for the variant present to be determined, one of which was a singleton introduction.
a, Frequency of amino acid residues at position 614 in the spike protein for Scottish sequences in March. D, aspartate; G, glycine; X, undetermined. b, Amino acid residue at spike 614 for each Scottish lineage introduction by date of first detection. c, Visualization of the genetic variation across the entire genome observed in the 1,314 SARS-CoV-2 genomes in Scotland. Non-synonymous (pink) and synonymous (green) substitutions (with respect to Wuhan-Hu-1; GenBank accession number MN908947) are represented in colour in each row. The mutations are plotted in a grid format where each row is a sample and each column is a unique genome position. Mutations have been filtered to only display those observed in more than two samples. Mutation labels have been added into the heatmap to show the genome position and open reading frame name, along with the amino acid number/substitution (non-synonymous mutation details are highlighted in bold). Labels for each of the open reading frames of the SARS-CoV-2 genome are shown in the bottom panel. The plot was created using the d3heatmap package in R, with samples (rows) ordered according to Ward’s clustering.
Introductions estimated based on phylogeny alone ranged from 234 (for the Scottish lineage as determined by the delayed transformation (DELTRAN) algorithm (del_sct_lineage)) to 1,035 (for Scottish phylogenetic introduction as determined by the DELTRAN algorithm (del_sct_introduction)) if no attempt was made to merge sibling lineages at the polytomies. We combined the phylogeny, travel history and date of sampling in order to refine these estimates. Merging sibling lineages provided a conservative phylogenetic estimate of introductions but failed to identify lineages comprising multiple introductions, of which there were 17 examples within our dataset (Supplementary Data 1). Conversely, failure to merge sibling lineages resulted in the inference of a large number of introductions that were unsupported by the available epidemiological data (for example, seven individuals with no travel history who were known to have been infected through a superspreader event at a conference were counted as seven separate introductions when the unmerged phylogenetic algorithm was applied) (Extended Data Fig. 4). Combining the more conservative del_sct_lineage estimate with the travel and temporal history permitted the confident identification of 283 independent introductions of SARS-CoV-2 into Scotland. In the majority of cases, the phylogenetic lineage placement correlated well with the available epidemiological information for introduction events. For example, all cases from two known superspreader events clustered together appropriately within single lineages, as did eight out of 12 known household contacts. Interestingly, two household contacts (EDB006 and EDB035, where ‘EDB’ represents analysis at the Virology Department, Royal Infirmary of Edinburgh) with common exposure history through travel to Italy in early March had differing lineage assignments, suggesting the acquisition of separate variants while travelling in a high-prevalence area at the time of exposure. A second pair of household contacts who had not travelled also had differing lineage assignments (EDB032 and EDB041) associated with three unique single-nucleotide polymorphisms, again suggesting an alternative source of infection.
Many of the introductions of COVID-19 into Scotland were from known returning travellers from Europe, mostly Italy (41 out of 283 introductions). Of 234 phylogenetic lineages, 140 (59.8%) were single cases not linked with further cases over time. Of these singleton lineages, 108 were not associated with travel, probably corresponding to undetected introductions and community transmission clusters. There were 94 phylogenetic lineages of at least two individuals associated with transmission in varied community settings (Fig. 4a–c). These included cases in the Shetland Islands (with travel links to Italy), cases associated with a care home facility, cases of community transmission across central Scotland and cases of transmission related to an international conference event in Edinburgh at the end of February before the first documented SARS-CoV-2 case in Scotland (Extended Data Fig. 4). The latter demonstrates the extent to which conferences and other large gatherings act as superspreader events and contribute to intensified spreading of the virus, supporting findings from similar events in China and Singapore18,19. Time-scaled trees were constructed in order to estimate the timing of undetected introductions in Scotland in lineages with no associated epidemiological travel history (Fig. 5). These inferred that the first introductions dated back as far as 19 February 2020, indicating that community transmission was likely to have occurred undetected up to 1–2 weeks earlier than the first detected cases. This is in keeping with previously published seroprevalence data from the Scottish Blood Transfusion Service20.
a, A spatially distributed outbreak across the Central Scotland belt without any known links to travel. b, A cluster focused on Shetland associated with two linked cases with travel to Italy. c, A focal outbreak in a residential facility not associated with any known travel. Labels include country of origin, sequence name, year, region and date of sample.
Time-scaled trees were produced with treetime (with a clock rate of 0.001 and ten iterations) for seven established lineages to give an indication of the detection lag (that is, the time between the most recent common ancestor and the first sequenced sample). Scottish lineage numbers are prefixed by SCT and equivalent UK lineages are shown in brackets.
Shift to community transmission
Ninety introductions (32%) were linked epidemiologically to travel in Europe, seven (2.5%) were linked to Caribbean cruises and seven (2.5%) were linked to travel to the rest of the world. The first case of documented community transmission occurred on 2 March 2020 and community transmission was well established by 11 March (Figs. 1 and 4a–c). Figure 4a represents a large cluster that spread across the Scottish central belt between 13 and 31 March with no known associated travel. A cluster occurring between 6 and 30 March that was focused on the Shetland Islands contained two index cases with a travel history linked to Italy (Fig. 4b), but all subsequent cases in the cluster were in individuals who did not report travel. This shift to community transmission is evident in other example clusters: Fig. 4c represents a cluster from a care home. We also investigated the possibility of local transmission chains in healthcare settings. As described above, healthcare workers were noted to represent some of the earliest cases, with an increase in cases over time (Extended Data Fig. 2). However, at the beginning of the outbreak, not all cases were community acquired and it was possible to exclude cases of potential nosocomial transmission by comparing sequences from patients on the ward with that of the healthcare worker. For example, a healthcare worker, CVR10 (Supplementary Data 1), showed evidence of infection from a virus strain distinct from other samples from the same hospital within lineage B.2.2, while the ward patient sequences were from lineages B.1.5 (CVR76), B.2.1 (CVR07) and B.1.10 (CVR79), indicating community rather than nosocomial infection. The increase in cases over time is likely to reflect a combination of increased testing in healthcare workers and nosocomial infection. Further studies of genetic epidemiology in healthcare settings are indicated.
Discussion
Our study indicates that SARS-CoV-2 entered the Scottish population through at least 283 separate travel-related introductions. This estimate was calculated using a combination of phylogenetic lineage and epidemiological data. In isolation, each dataset would have resulted in an underestimate of the number of introductions due to the slow rate of evolution of the SARS-CoV-2 virus and the absence of detection of travel-related incident cases, respectively. There were 234 phylogenetic lineages, of which 94 (40%) were associated with sustained community transmission (containing at least two Scottish sequences without a history of travel) and 140 (60%) were singleton sequences with no evidence of onward transmission. Of the 94 phylogenetic lineages associated with onward transmission, 34 (36%) involved individuals with a known history of international travel. The majority returned to Scotland from Europe at the end of February and early March following travel to Italy and, less commonly, to Spain, Austria, Switzerland, France, England, Ireland, Poland, the Caribbean and Thailand. While the first positive case occurred on 1 March 2020, evidence of community transmission during late February 2020 was supported by epidemiological data and by time-scaled phylogenetic analysis. A shift from travel-associated infection in younger adults to community transmission in older adults and healthcare workers was noted throughout the first month of the epidemic.
On 28 January 2020, the UK Government recommended against all but essential travel to China and for returning travellers to self-isolate for 2 weeks upon their return, regardless of symptoms21. However, in this study, cases with direct links to Southeast Asia were rare (only one case associated with travel to Thailand was detected through epidemiological analysis). In contrast, travel to continental Europe in February and March 2020 (by then the epicentre of the global COVID-19 pandemic) was a clear driver of the Scottish outbreak; the majority of the lineages detected in this study were lineage B and related to European sequences. Lineage A—a lineage with a distribution more limited to China at the beginning of the outbreak—was introduced to Scotland on at least ten occasions. The travel history and phylogenetic analysis of these cases indicated that the majority of these occurred via Spain. One introduction was potentially attributable to importation from China (and one from the United States) based on phylogenetic evidence alone. Despite evidence of local transmission in Italy as early as 21 February, the advice from the Scottish Government for returning travellers from Italy to self-isolate was issued only on 25 February and was limited to those having returned from specific lockdown areas22,23,24. By the time this advice was extended to all travellers on 10 March, the COVID-19 outbreak within Scotland was already being driven by community transmission. A lack of robust measures to manage ingress of high numbers of infected travellers from rapidly emerging pandemic hotspots may have accelerated the course of the outbreak in Scotland and the United Kingdom as a whole.
Our data demonstrate that SARS-CoV-2 was introduced to Scotland on many hundreds of occasions, so no single event can be considered to have sparked the epidemic in the rest of the country. One notable introduction event at the beginning of the outbreak occurred at an international conference held in Edinburgh during late February, several days before the first Scottish case was confirmed. Several cases were linked to this event, with the last case within the cluster in the United Kingdom occurring on 27 March, showing that the local public health response was effective in controlling the spread. However, the geographical distribution of related sequences is striking, spanning four continents and ten countries. The role of this event in local dispersal of the virus, before a single case had been identified in Scotland, demonstrates that governments should be wary of prematurely relaxing restrictions on large gatherings and international travel. Importantly, the directionality of transmission cannot be inferred from the phylogeny; SARS-CoV-2 could have been introduced from the Netherlands and other countries to Scotland, exported from Scotland or both.
In parallel with the above introduction, we identified other viral lineages with no epidemiological link to travel as early as 3 d after the first detection of infection, suggesting earlier introduction to Scotland than the first detected case, which was reported on 1 March 202015. This is supported by the analysis of time-scaled phylogenies, which infers a common ancestor from mid- to late February for some Scotland-specific clades. The majority of individuals had no recorded link to travel by 12 March, only 2 d after substantial travel and physical distancing restrictions were put in place. Importantly, these data are suggestive of introductions and community spread well before initial detection. The epidemic in the United Kingdom expanded rapidly, prompting the government to respond with restrictive public health measures or lockdown to disrupt transmission. These phylogenetic data will provide a baseline for granular real-time sequencing of infections as cases rise and fall over time and will be used as a measure of the success of current measures with the potential to contribute to decision-making around the easing (or tightening) of public health measures. In this study, the use of epidemiology or phylogenetic lineage assignment alone would have resulted in an underestimate of the number of introductions of the virus (114 and 234, respectively) due to undetected cases reaching the attention of public health authorities and due to the slow rate of evolution of the virus. Integration of genomic sequence data with traditional case finding and contact tracing has the potential to enhance descriptive epidemiology and deliver more targeted control measures. However, previous interventions of this type have been largely retrospective. The challenge will be to develop a framework where phylogenetic information is delivered in real time in an easily actionable format to public health and infection control teams.
This study has some limitations. We sampled only around half of positive samples detected during the initial outbreak; therefore, some introduction events will have been missed. Furthermore, while laboratory and hospital case notes were available for review, public health records were not accessed to record linkages with sequence data. Our analysis is likely to have been affected by a shift from sampling all symptomatic individuals to hospitalized patients and healthcare workers only during mid-March. Some travel-related introductions may therefore not have been detected (as evidenced by several clusters with no evidence of travel-related infection after this time) and healthcare workers may have been over-sampled in the analysis. While the introduction of erroneous variation during sequencing or genome assembly is possible, we estimated experimentally that such rates were extremely low and should not affect clustering patterns in phylogenetic analyses. This analysis, based on the available phylogeny, is most likely to be an underestimate, despite dense sampling of the beginning of the outbreak, as many events may be linked to an identical sequence due to the slow evolution of SARS-CoV-2. Several events that we counted as introductions were based on epidemiological history but could not be resolved by phylogeny alone. The slow evolutionary rate of the virus also means that linkage of sequences does not categorically prove that transmission events have occurred and require correlation with epidemiological information. Exclusion of linkage may be inferred with more certainty. Nevertheless, as the variation present within the global diversity is well represented in Scotland, reflecting a high number of near-simultaneous introductions, tracking of the outbreak is feasible and can be used to refine public health interventions. Finally, as further sequence data become available from other countries, the topology of the global tree is predicted to change, which is likely to have the effect of a slight increase in our estimate of introductions.
In summary, the first month of the COVID-19 outbreak in Scotland was associated with multiple introductions related to travellers returning from Europe, early community transmission and clusters related to large indoor events and healthcare facilities. Earlier lockdown in countries with a high burden of cases, such as Italy, and other measures such as quarantine of travellers returning from high-risk areas might have prevented escalation of the outbreak and multiple clusters of ongoing community transmission. Combining genomic data with epidemiological data has the potential to inform public health intervention policy. Multiple travel-associated introductions during the first wave of infection in Scotland highlight this as a key risk factor for reintroduction in future waves.
Methods
Samples
Up to 300 samples per week were selected prospectively following ethical approval from the relevant national biorepository authorities covering the sequencing laboratories (16/WS/0207NHS (Glasgow) and 10/S1402/33 (Edinburgh)) between 1 March and 1 April 2020. This work was conducted as part of the UK Government-funded COVID-19 Genomics (COG-UK) Consortium, which was set up in March 2020 and aims to provide representative, large-scale and rapid whole-genome virus sequencing across the United Kingdom25. Half of the samples were randomly selected to achieve a representative target for all Scottish health boards and the other half to cover suspected healthcare-related nosocomial infections as they occurred. Health boards with a small population size were reported at a minimum of five sequences per region to avoid deductive disclosure. The Royal Infirmary of Edinburgh (RIE) and West of Scotland Specialist Virology Centre, NHS Greater Glasgow and Clyde conducted diagnostic real-time PCR with reverse transcription to detect SARS-CoV-2-positive samples following nucleic acid extraction utilizing the NucliSENS easyMAG and Roche MG96 platforms26. Residual nucleic acid from 1,314 samples underwent whole-genome next-generation sequencing at the CVR and RIE. Clinical details, including recent travel history, were obtained from assay request forms submitted to the diagnostic laboratory and, where available, electronic patient records and local public health databases. A travel history was defined as travel from any country (including England, Wales and Northern Ireland) by any means of transport (including by air, ferry, train or car) within the 2 weeks before the onset of symptoms. Statistical comparisons (two tailed) were carried out using R version 3.6.3 by Kruskal–Wallis test.
Rapid sequencing protocol using Oxford Nanopore Technologies
Following extraction, libraries were prepared utilizing protocols developed by the ARTIC Network (versions 1 and 2; https://artic.network/ncov-2019). Library pools (50 fmol) were loaded onto each flow cell (R9.4.1). Sequencing was conducted in MinKNOW version 19.12.5. Raw FAST5 files were base called using Guppy version 3.4.5 in high accuracy mode using a minimum quality score of 7. RAMPART version 1.0.5 was used to visualize the read mapping in real time. Reads were size filtered, demultiplexed and trimmed with Porechop version 0.2.4 (https://github.com/rrwick/Porechop) and mapped against the reference strain Wuhan-Hu-1 (MN908947). Variants were called using Nanopolish 0.11.3 and accepted if they had a log-likelihood score of greater than 200 and a minimum read coverage of 20 following the ARTIC bioinformatics pipeline version 1.1.3 (https://github.com/artic-network/artic-ncov2019). This protocol was used by the CVR and RIE sites.
High-throughput sequencing protocol using Illumina MiSeq
Amplicons were generated as described above. DNA fragments were cleaned using AMPure beads (Beckman Coulter) and 40 ng were used to prepare Illumina sequencing libraries with a DNA KAPA Library Preparation Kit (Roche). Indexing was carried out with NEBNext Multiplex Oligos (New England Biolabs) using seven cycles of PCR. Libraries were pooled in equimolar amounts and loaded on a MiSeq V2 cartridge (500 cycles). Reads were trimmed with Trim Galore version 0.6.5 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and mapped with Burrows–Wheeler Aligner27 to the Wuhan-Hu-1 (MN908947) reference sequence, followed by primer trimming and consensus calling with iVar version 1.2.2 (ref. 28) and a minimum read coverage of ten. This protocol was followed by the CVR site, whereas samples sequenced at the Wellcome Sanger Institute (labelled with the prefix GCVR) followed a diverse Illumina library preparation method and used the NovaSeq instrument. Consensus sequence data were highly correlated between different sequencing platforms.
Sequence data
Consensus sequences with >90% coverage were included. All consensus genomes are available from the GISAID database (https://www.gisaid.org), the COG-UK Consortium website (https://www.cogconsortium.uk/data/) and BAM files from the European Nucleotide Archive’s Sequence Read Archive service (with BioProject ID PRJEB37886). See the source data for IDs and dates of sampling. Amplicon-based sequencing using Illumina instruments has previously been applied to RNA viruses for accurate identification of intra-host variants, with performance comparable to metagenomics28. A similar protocol has been applied to dengue virus using multiplex PCR tiling on an Oxford Nanopore sequencer, resulting in 99.69–99.92% consensus identity when compared with those sequences produced by Illumina29. To minimize consensus-level errors in regions of lower genome coverage and in samples with very low viral load28, we opted to exclude samples with poor genome coverage (<90%; typically associated with low viral load) while: (1) sequencing at a high depth with an average of 5,000× per sample (CVR); or (2) actively excluding samples with a low viral load (Ct > 30) while sequencing at an average depth of 500× (RIE).
Phylogenetic analysis
The grapevine pipeline (https://github.com/COG-UK/grapevine) was used for generating the phylogeny based on all data available on GISAID and COG-UK up until 23 August 2020. Briefly, the pipeline cleans and filters reads based on quality and consensus coverage, aligns the sequences to the reference, types the sequences using PANGOLIN version 2.0.5 (https://github.com/hCoV-2019/pangolin), merges the COG-UK sequences with the GISAID alignment, masks homoplasies (position 11,083 relative to Wuhan-Hu-1) and reconstructs the maximum-likelihood phylogeny using FastTree version 2.1.11 (ref. 30). Wuhan/WH04/2020 was used as an outgroup for the phylogeny. Finally, the phylogeny was pruned to keep only sequences before 31 March 2020, to retrospectively investigate the introductions into Scotland alongside known travel history and epidemiological information. Scottish clusters were investigated following the DELTRAN implementation method31 with the following four steps using clusterfunk version 0.0.3 (https://github.com/cov-ert/clusterfunk): (1) Scottish and non-Scottish sequences were coded as a binary trait and annotated on the tree; (2) the ancestral state of the Scottish trait was reconstructed on the phylogeny; (3) transition nodes from non-Scottish to Scottish were identified as DELTRAN Scottish introductions on the tree (del_sct_introductions); and (4) transitions were then merged such that sibling introductions were clustered together, ensuring that identical sequences were given the same DELTRAN Scottish lineage number (del_sct_lineage). This was a fully automated approach with some caveats. First, due to the slow evolutionary rate of the virus, the tree has many polytomies and thus the parsimony reconstruction of a trait on the tree becomes ambiguous (hence the need to merge sibling introductions together). Additionally, where there are exports and subsequent re-introductions (unlikely during the first month of the outbreak), the DELTRAN approach would label these as the ancestral introduction.
Due to the relatively low evolutionary rate of SARS-CoV-2, it is often difficult to differentiate between introductions on the basis of the phylogeny alone, as sequences from distinct introductions may be identical or cluster on the phylogeny (thereby underestimating introductions), and incorporation of the assumption that identical sequences at the base of the tree represent distinct introductions may result in an overestimate. To refine the phylogenetic estimate of the number of times the virus was introduced into Scotland, we used the more conservative del_sct_lineage as a framework on which we layered available epidemiological and temporal data. Lineages with no known travel-related cases were counted as a single introduction regardless of the number of taxa present. Multiple travel-related cases within a lineage were each classed as separate introductions. Where a single travel-related case was recorded within a phylogenetic lineage, this was counted as a single introduction if it occurred up to 14 d after the first case in the cluster. If the travel-related case returned to Scotland after the lineage had already been detected, or if the travel-related case was sampled more than 14 d following the first detection of the lineage within Scotland, it was counted as a further introduction.
Seven large UK lineages that were predominantly sampled from Scotland were investigated further using time-scaled trees with treetime32 (with a clock rate of 0.001 and ten iterations) in order to give an indication of the detection lag (that is, the time between the most recent common ancestor and the first sequenced sample).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All consensus genomes are available from the GISAID database (https://www.gisaid.org), the COG-UK Consortium website (https://www.cogconsortium.uk/data/) and BAM files from the European Nucleotide Archive’s Sequence Read Archive service (BioProject ID: PRJEB37886). Source data are provided with this paper.
Code availability
Reads were size filtered, demultiplexed and trimmed with Porechop version 0.2.4 (https://github.com/rrwick/Porechop). The ARTIC bioinformatics pipeline version 1.1.3 (https://github.com/artic-network/artic-ncov2019) was used for the generation of consensus sequences. The grapevine pipeline (https://github.com/COG-UK/grapevine) was used to generate the phylogeny based on all available data on GISAID and COG-UK up until 23 August 2020. Sequence typing was carried out using PANGOLIN version 2.0.5 (https://github.com/hCoV-2019/pangolin). Scottish clusters were investigated following the DELTRAN implementation method using clusterfunk version 0.0.3 (https://github.com/cov-ert/clusterfunk).
Change history
18 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41564-021-00865-4.
27 January 2021
A Correction to this paper has been published: <ExternalRef><RefSource>https://doi.org/10.1038/s41564-021-00869-0</RefSource><RefTarget Address="10.1038/s41564-021-00869-0" TargetType="DOI"/></ExternalRef>.
References
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Mao, L. et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 77, 683–690 (2020).
Verdoni, L. et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet 395, 1771–1778 (2020).
Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV) (World Health Organization, 2020).
WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020 (World Health Organization, 2020).
Quick, J. et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016).
Grubaugh, N. D. et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546, 401–405 (2017).
Grubaugh, N. D. et al. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 4, 10–19 (2019).
Kafetzopoulou, L. E. et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science 363, 74–77 (2019).
Biek, R., Pybus, O. G., Lloyd-Smith, J. O. & Didelot, X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 30, 306–313 (2015).
Hill, K. J. et al. The index case of SARS-CoV-2 in Scotland. J. Infect. 81, 147–178 (2020).
Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020).
Korber, B. et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827 (2020).
Tong, Z. D. et al. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg. Infect. Dis. 26, 1052–1054 (2020).
Pung, R. et al. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet 395, 1039–1046 (2020).
Thompson, C. P. et al. Detection of neutralising antibodies to SARS coronavirus 2 to determine population exposure in Scottish blood donors between March and May 2020. Euro Surveill. 25, 2000685 (2020).
Foreign Office Advises Against All but Essential Travel to China (Foreign and Commonwealth Office, 2020); https://www.gov.uk/government/news/fco-advises-against-all-but-essential-travel-to-mainland-china
Preparations for Coronavirus Stepped Up (Scottish Government, 2020); https://www.gov.scot/news/preparations-for-coronavirus-stepped-up
Communicable Disease Threats Report; 16–22 February 2020, Week 8 (European Centre for Disease Prevention and Control, 2020).
Outbreak of Novel Coronavirus Disease 2019 (COVID19): Situation in Italy—23 February 2020 (European Centre for Disease Prevention and Control, 2020).
The COVID-19 Genomics UK (COG-UK) Consortium. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe 1, e99–e100 (2020).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Grubaugh, N. D. et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 20, 8 (2019).
Stubbs, S. C. B. et al. Assessment of a multiplex PCR and nanopore-based method for dengue virus sequencing in Indonesia. Virol. J. 17, 24 (2020).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Farris, J. S. Methods for computing Wagner trees. Syst. Zool. 19, 83–92 (1970).
Sagulenko, P., Puller, V. & Neher, R. A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 4, vex042 (2018).
Acknowledgements
We thank all staff at the Scottish NHS virology laboratories who provided samples for sequencing, as well as the global researchers who shared genome data on GISAID (https://www.gisaid.org). We thank R. Kuo, T. Regan and A. Warr (The Roslin Institute, University of Edinburgh) for sequencing reagents, and S. Arkison for server maintenance. The study was funded by the Medical Research Council (MRC) core award (MC UU 1201412), a Wellcome Trust Collaborator Award (206298/Z/17/Z – for the ARTIC Network), and a Wellcome Trust Award (204802/Z/16/Z). COG-UK is supported by funding from the MRC part of UK Research and Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute. The Cloud Infrastructure for Microbial Bioinformatics is funded by the MRC through grant MR/L015080/1.
Author information
Authors and Affiliations
Consortia
Contributions
C.B., R.D., M.D.G., M.P.M., S.R., E.W., K.T., B.J.P., E.J., K.N., R.G., I.S., A.M., K.L., J.S. and N. Johnson acquired the clinical samples and performed extractions and quantitative PCR. T.W., R.D., M.P.M., K.T., B.J.P., E.J., K.N., P.A., S.A., C.D., M.N., S.E.M., R.S., K.L., J.S., N. Jesudason and E.C.T. were responsible for the selection, reception, verification and organization of clinical samples. J.G.S., T.W., E.W., A.C., R.G., A.H., E.J., N. Jesudason, K.L., D.A.M., K.M., B.J.P., R.S., I.S., M.T.G.H., K.T., A.K.M. and E.C.T. collected and analysed the clinical and epidemiological data. T.W., R.D., M.D.G., V.H., B.J., J.T.M., M.P.M., A.O., A.R., S.R., K.T., E.A.-C., K.B., S.C., N. Johnson, E.C.M.L., D.M., J.N., K.N., Y.A.P., K.S., L.T. and A.d.S.F. acquired the sequencing data and optimized protocols. M.D.G., V.H., B.J., J.T.M., A.O., A.R., S.R., J.H., R.J.O., V.B.S. and D.L.R. were responsible for the genome alignment and variant calling. J.H., J.B.S., E.C.T., J.S. and D.L.R. performed the phylogenetic analysis, integration of epidemiology analysis and genome mutation analysis. A.d.S.F., J.S., T.W., J.H., D.L.R., M.T.G.H., S.S., T.W., K.T., M.P. and E.C.T. analysed and interpreted the data. A.d.S.F., J.S., T.W., J.H., D.L.R. and E.C.T. drafted the manuscript. All authors reviewed and contributed to the final version of the manuscript. A.d.S.F., J.S., T.W., J.H., D.L.R., M.P., K.T. and E.C.T. were responsible for the conception and design of the study. M.P., E.C.T. and A.R. acquired funds to support the study. All authors approved the submitted version and have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated and resolved, and the resolution documented in the literature.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Damien Tully and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Geographical sequencing coverage in Scotland for March 2020.
A; Scottish population density (population/km2) by council area, B; number of laboratory-confirmed cases of COVID-19 by council area C; proportion of laboratory confirmed cases sequenced by council area. Population and testing data obtained from https://datashare.is.ed.ac.uk/handle/10283/2558 and https://www.opendata.nhs.scot/dataset/covid-19-in-scotland.
Extended Data Fig. 2 Number of laboratory-confirmed cases of COVID-19 in each of the first four weeks of the outbreak in Scotland.
The contribution of healthcare workers is coloured in orange. The percentage of HCW testing positive in each week is indicated in orange text.
Extended Data Fig. 3 Lineage and travel destination of sequenced individuals.
Relative contribution of each PANGOLIN lineage (upper panel), and travel destination of sequenced individuals (lower panel) by date of detection for sequences sampled in Scotland during March 2020.
Extended Data Fig. 4 A phylogenetic cluster associated with an international event hosted in Scotland in February 2020.
This cluster was associated with international dispersal of this sub-lineage as indicated.
Supplementary information
Supplementary Information
Members of the COG-UK Consortium.
Supplementary Data 1
Phylogenetic tree showing the relationship of the 1,314 Scottish genomes to global SARS-CoV-2 genomes sampled prior to April 2020. Scottish sequences are annotated in the form ‘Scotland|postcode district|sequence name_sampling date|travel history|del_sct_lineage’. Non-Scottish sequences are annotated in the form ‘sampling location|sequence name_sampling date’. Scottish tip shapes are coloured by UK residential postcode area and tip labels coloured blue. The scale bar indicates substitutions per nucleotide site.
Source data
Source Data Fig. 1
Source data used for Figs. 1–5 and Supplementary Data 1. Associated metadata. Column headers are as follows: COG-UK sequence ID (sequence_name); GISAID sequence ID (gisaid_id); GISAID accession number (gisaid_accession); date of submission to GISAID (gisaid_submission_date); date of sample collection (sample_date); pangolin lineage assignment (pangolin); amino acid residue at position 614 in spike (spike_614); amino acid residue at position 439 in spike (spike_439); UK lineage as determined by the DELTRAN algorithm (uk_lineage); Scottish phylogenetic introduction as determined by the DELTRAN algorithm (del_sct_introduction), Scottish lineage as determined by the DELTRAN algorithm (del_sct_lineage), history and location of travel in the 2 weeks before symptom onset (travel_hx); Scottish introduction as determined by the combination of del_sct_lineage and epidemiological information (combined_sct_intro); and the sequencing platform used to generate the viral sequence (seq_technology).
Source Data Fig. 2
Source genetic data used for phylogenetic analysis and affiliations of contributors, associated with Figs. 1–5 and Supplementary Data 1.
Rights and permissions
About this article
Cite this article
da Silva Filipe, A., Shepherd, J.G., Williams, T. et al. Genomic epidemiology reveals multiple introductions of SARS-CoV-2 from mainland Europe into Scotland. Nat Microbiol 6, 112–122 (2021). https://doi.org/10.1038/s41564-020-00838-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-00838-z
This article is cited by
-
Evolution of enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants
Nature Microbiology (2024)
-
Maximum likelihood pandemic-scale phylogenetics
Nature Genetics (2023)
-
An appeal for strengthening genomic pathogen surveillance to improve pandemic preparedness and infection prevention: the German perspective
Infection (2023)
-
Public health genomics capacity assessment: readiness for large-scale pathogen genomic surveillance in Canada’s public health laboratories
BMC Public Health (2022)
-
Ecological and evolutionary dynamics of multi-strain RNA viruses
Nature Ecology & Evolution (2022)